Introduction
Clinical trials for using hematopoietic stem cell transplant (HSCT) with gene therapies to cure sickle cell disease (SCD) began in 2019. The Food and Drug Administration (FDA) requires that 1) all gene therapy participants receive at least 15 years of ongoing surveillance and 2) their outcomes be compared to a contemporaneous cohort that did not receive gene therapy. Previously we developed an automated contemporaneous cohort of children and adults with SCD from electronic health record (EHR) data (Cronin RM et al. 2023, Blood Adv). Our work was built upon previously explored algorithms utilizing ICD codes and laboratory data to identify persons with sickle cell anemia (SCA), a severe form of SCD (Singh et al. 2018, Blood Adv). Still, our algorithm additionally classifies other predicted genotypes. However, we could not test the SCD phenotypes against the SCD genotypes due to the absence of beta-globin gene sequence data. Here we tested the hypothesis that our Vanderbilt-EHR algorithm for identifying a contemporaneous cohort at Vanderbilt can classify their predicted genotype compared to beta-globin gene sequencing, the gold standard for SCD diagnosis, in the cohort. We subsequently tested the Vanderbilt-EHR algorithm in an established cohort of children from St. Jude Children's Research Hospital that have been deeply phenotyped.
Methods
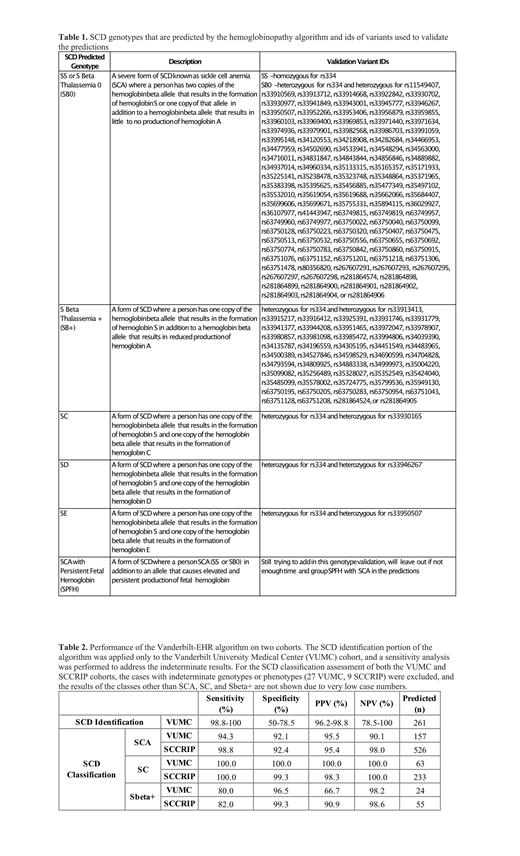
We applied the Vanderbilt-EHR algorithm to a cohort of 275 samples from Vanderbilt's biorepository of DNA, BioVU, linked to de-identified medical records in a data warehouse called the Synthetic Derivative. These 275 samples were previously submitted for whole genome sequencing as part of the Genetic Variation of Heart, Lung, and Kidney Disease in Sickle Cell Disease: Pre- and Post-Curative Therapies TOPMed project (HLK-SCD phs002617). The returned genotypes were validated using the hemoglobin variant database HbVar (Giardine BM et al. 2021, Nucleic Acids Res) (Table 1). We applied the hemoglobinopathy genotype classification portion of the algorithm to the Sickle Cell Clinical Research and Intervention Program (SCCRIP) cohort from St. Jude Children's Research Hospital, which includes putative genotype diagnoses derived from a comprehensive evaluation of clinical data for each member. We performed statistical analyses of the identification and classification results of the Vanderbilt cohort and the SCCRIP cohort's classification results.
Results
Of the 275 genotyped samples, the algorithm correctly predicted 255 SCD cases and 10 non-SCD cases. There were three cases predicted to be false positives; however, a comprehensive analysis showed that all have laboratory values indicative of SCA (high hemoglobin S levels >60%, absent or negligible hemoglobin A levels, elevated reticulocytes, and low mean corpuscular volume) suggesting that there may be a genotyping error. The remaining 7 cases were either indeterminate in the prediction or in the genotype (Table 1). Given the small number of indeterminate results, we performed a sensitivity analysis to identify the range of performance for SCD identification. The method performed very well with over 98% sensitivity and over 96% positive predictive value (PPV) (Table 2). The lower specificity and negative predictive value (NPV) could be largely attributed to the disproportionately high number of SCD cases in the dataset compared to non-SCD cases. The SCD classification algorithm performed similarly in both the VUMC and SCCRIP cohorts (Table 2). The algorithm performs best in identifying SC and SCA types of SCD with nearly 100% accuracy for SC and over 94% accuracy for SCA. Transfusions can confound the diagnosis of SCA vs. S beta thalassemia +, but the latter has a much lower prevalence consistent with our results (Table 2).
Discussion and Conclusions
Larger SCD cohorts will be identified using these approaches in singular EHR databases and in de-identified data warehouses that EHR companies have developed by aggregating data across multiple EHR systems (e.g., EPIC Cosmos and Cerner Real-World Data). The ongoing need for contemporaneous comparison cohorts of persons with SCD to understand outcomes related to treatment and phenotype can be greatly assisted by accurate automated approaches. The Vanderbilt-EHR algorithm is the first to comprehensively phenotype SCD and other hemoglobinopathies involving variant hemoglobin beta alleles.
Disclosures
Saraf:Agios: Consultancy, Other: Advisory board; BEAM Therapeutics: Consultancy, Other: Advisory board; Novartis: Consultancy, Other: Advisory board, Research Funding; Forma Therapeutics: Consultancy, Other: Advisory board, Research Funding; GBT/Pfizer: Consultancy, Other: Advisory board, Research Funding, Speakers Bureau. Weiss:Forma Therapeutics: Consultancy; Dyne: Consultancy; Graphite Bio: Consultancy; Novartis: Consultancy; Cellarity Inc.: Consultancy. DeBaun:Novartis, Forma, Vertex: Consultancy, Other: Consulting.